How to Use Autodock Vina Online
Try Autodock Vina
Commercially Available Online Web Server
AutoDock Vina: Accelerating Molecular Docking and Virtual Screening
Molecular docking is a fundamental computational technique in drug discovery that predicts how a small molecule (a ligand) binds to a larger molecule like a protein (receptor). The goal is to predict the bound conformation and the binding affinity between the two molecules. This process is crucial for screening large virtual libraries of drug-like molecules to identify promising candidates for further development. AutoDock Vina is a popular, open-source program that has significantly improved the speed and accuracy of molecular docking. It's up to two orders of magnitude faster than its predecessor, AutoDock 4, while also improving the accuracy of its binding mode predictions.
How AutoDock Vina Works
AutoDock Vina is designed to be a "turnkey" program, meaning it requires minimal user input to perform a docking simulation. Unlike older programs that required pre-calculating grid maps for each atom type, Vina handles this automatically and transparently to the user. It also automatically clusters results, simplifying the analysis.
The program's core lies in its efficient optimization algorithm and scoring function.
Scoring Function: Vina uses an empirically derived scoring function that approximates the binding affinity. This function considers various factors like hydrogen bonding, hydrophobic interactions, and steric complementarity.
Search Algorithm: Vina employs a stochastic global optimization combined with a gradient-based approach to explore the conformational space of the ligand. It starts with random conformations and uses a local optimizer (a Broyden-Fletcher-Goldfarb-Shanno algorithm) to refine the pose. This process is repeated multiple times to find the most favorable binding poses. The use of multithreading on multi-core machines allows for parallel execution of these independent runs, which further speeds up the process.
Efficient Optimization: The software employs an efficient optimization method to search the conformational space of the molecules being docked.
Multithreading: Further speed-up is achieved through parallelism, by using multithreading on multi-core machines.
Automation: The program automatically calculates the necessary grid maps and clusters the results in a way that is transparent to the user, streamlining the entire workflow.
Performance: AutoDock Vina provides an approximate two orders of magnitude speed-up compared to AutoDock 4, while simultaneously improving the accuracy of binding mode predictions.
Comparing AutoDock Vina & smina
Smina represents a fork of the AutoDock Vina model. It is often important to compare both models.
AutoDock Vina | smina | |
|---|---|---|
Origin | The original, widely-used molecular docking program developed by Oleg Trott and Arthur Olson at The Scripps Research Institute. | A fork of AutoDock Vina, developed by David Koes' lab at the University of Pittsburgh. |
Primary Focus | General-purpose, fast, and easy-to-use docking for virtual screening and pose prediction. | Optimized for scoring function development and high-performance energy minimization. |
Scoring Function | Uses a default, built-in empirical scoring function based on a combination of Gaussian, repulsion, hydrophobic, and hydrogen-bonding terms. | Supports the default Vina scoring function but also allows for user-defined, custom scoring functions and additional terms like desolvation and electrostatics. |
Minimization | Provides a robust gradient-optimization conformational search. | Enhanced and faster energy minimization, making it particularly useful for workflows that focus on refining a known pose (local searches). |
Input/Output Formats | Primarily uses the PDBQT format for both receptor and ligand files. | Supports a wider range of molecular formats (e.g., SDF) in addition to PDBQT, which facilitates integration with other cheminformatics tools like OpenBabel. |
Flexibility | Allows for flexible side chains on the receptor and rotatable bonds in the ligand. | Similar flexible docking capabilities with some reported improvements in handling ligand and receptor flexibility, but with a focus on improving the scoring of flexible systems. |
Usability | Known for its simplicity and minimal required user input. It automatically handles many steps, such as grid map calculations. | While command-line driven like Vina, it offers more fine-grained control through additional parameters and switches. It's often preferred by advanced users and developers. |
Development | Maintained and developed by the Forli Lab at Scripps. | Maintained by David Koes and his team and is not officially part of the AutoDock suite. |
What is Tamarind Bio?
Tamarind Bio is a pioneering no-code bioinformatics platform built to democratize access to powerful computational tools for life scientists and researchers. Recognizing that many cutting-edge machine learning models are often difficult to deploy and use, Tamarind provides an intuitive, web-based environment that completely abstracts away the complexities of high-performance computing, software dependencies, and command-line interfaces.
The platform is designed provide easy access to biologists, chemists, and other researchers who may not have a background in programming or cloud infrastructure but want to run experimental models with their data. Key features include a user-friendly graphical interface for setting up and launching experiments, a robust API for integration into existing research pipelines, and an automated system for managing and scaling computational resources. By handling the technical heavy lifting, Tamarind empowers researchers to concentrate on their scientific questions and accelerate the pace of discovery. The Tamarind team hold information/data security as a top priority, as detailed in our Trust Center & Terms of Service, ensuring your data is safe on the platform.
The Role of Tamarind in Molecular Docking
Despite its ease of use, running large-scale docking simulations with AutoDock Vina, such as virtual screening of millions of compounds, still requires significant computational resources. Many researchers in academia and industry lack access to the necessary hardware and expertise to manage these complex workflows.
Tamarind is a no-code bioinformatics platform that makes advanced computational tools like AutoDock Vina readily available to scientists. It offers a simple web interface and API that handles the computational heavy lifting, including:
Massive-Scale Virtual Screening: Tamarind's infrastructure allows researchers to perform high-throughput virtual screening with AutoDock Vina on hundreds of thousands of compounds simultaneously, a task that would be infeasible on a single machine.
Pipeline Integration: The platform allows for the creation of seamless pipelines. For example, a researcher could start with a protein structure, use AutoDock Vina to screen a library of potential ligands, and then use other tools like DiffDock or molecular dynamics simulations to further analyze the top hits and assess binding affinity.
Simplified Workflows: Researchers don't need to manually prepare input files or manage the docking process. The platform automates these steps, freeing up scientists to focus on interpreting results and designing their next experiments.
Accurate Prediction: The improved accuracy of Vina's binding mode predictions increases the reliability of virtual screening, leading to higher-quality drug candidates for experimental validation.
Optimized Performance: By leveraging cloud infrastructure and GPU orchestration, Tamarind can run Vina simulations at a speed and scale that is not possible with traditional local setups.
How to Use AutoDock Vina on Tamarind Bio
To leverage AutoDock Vina's power, a researcher could follow this streamlined workflow on Tamarind:
Access the Platform: Begin by logging in to the tamarind.bio website.
Select AutoDock Vina: From the list of available computational models, choose the AutoDock Vina tool.
Input Structures: Provide the 3D structures of the protein (receptor) and the small molecule(s) (ligand) you wish to dock.
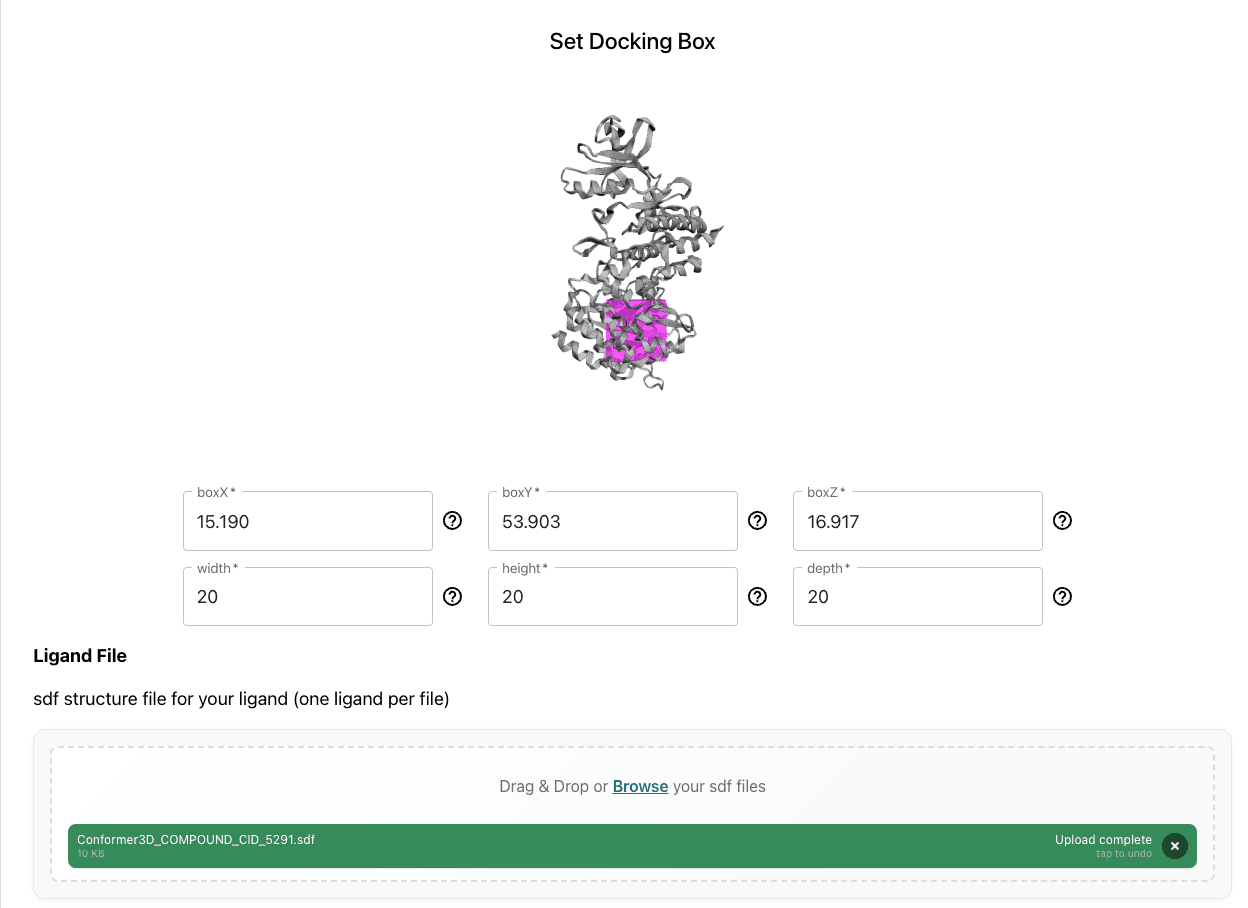
Define the Search Space: Define the search space around the expected binding site on the receptor structure.
Run Docking: The platform would automatically handle the parallelization and grid map calculations.
Analyze Results: The tool provides the predicted binding modes and scores, which are clustered for easy analysis, allowing you to quickly identify the best ligand candidates.